SuCComBase Manual
- Site Organization
- The Compounds table contains all the SCC compounds identified from literature and KNApSAcK database. SCC compounds information specifically produced by Arabidopsis thaliana, Carica papaya, Brassica rapa, and Brassica oleracea were collected.
- SCC genes table listed all the known Arabidopsis genes to involve in SCC biosynthesis through experiments.
- Putative genes table consists of all putative Arabidopsis SCC genes which computationally predicted from metabolic pathway databases, Kyoto Encyclopedia of Genes and Genomes (KEGG) and AraCyc.
- Potential genes table listed all the genes that co-expressed with the known SCC genes. The co-expression network data were obtained from three public databases, AraNet, GeneMania and ATTED.
- SCC homologs table contains SCC genes identified in other three Brassicales plants. All SCC homologs were collected from the sequence homology searching activity against three plant genomes, Carica papaya, Brassica rapa, and Brassica oleracea using Phytozome database Blast tool.
- Publications table comprised all the publications related to SCC biosynthetic genes in Brassicales.
- Search Box
- AGI ID
- Uniprot ID
- Refseq ID
- Gene name
- Compound name or common name
- Compound formula
- Knapsack ID
- Keyword, e.g. glucosinolate
- Browse
- Datasets
- Entry Details
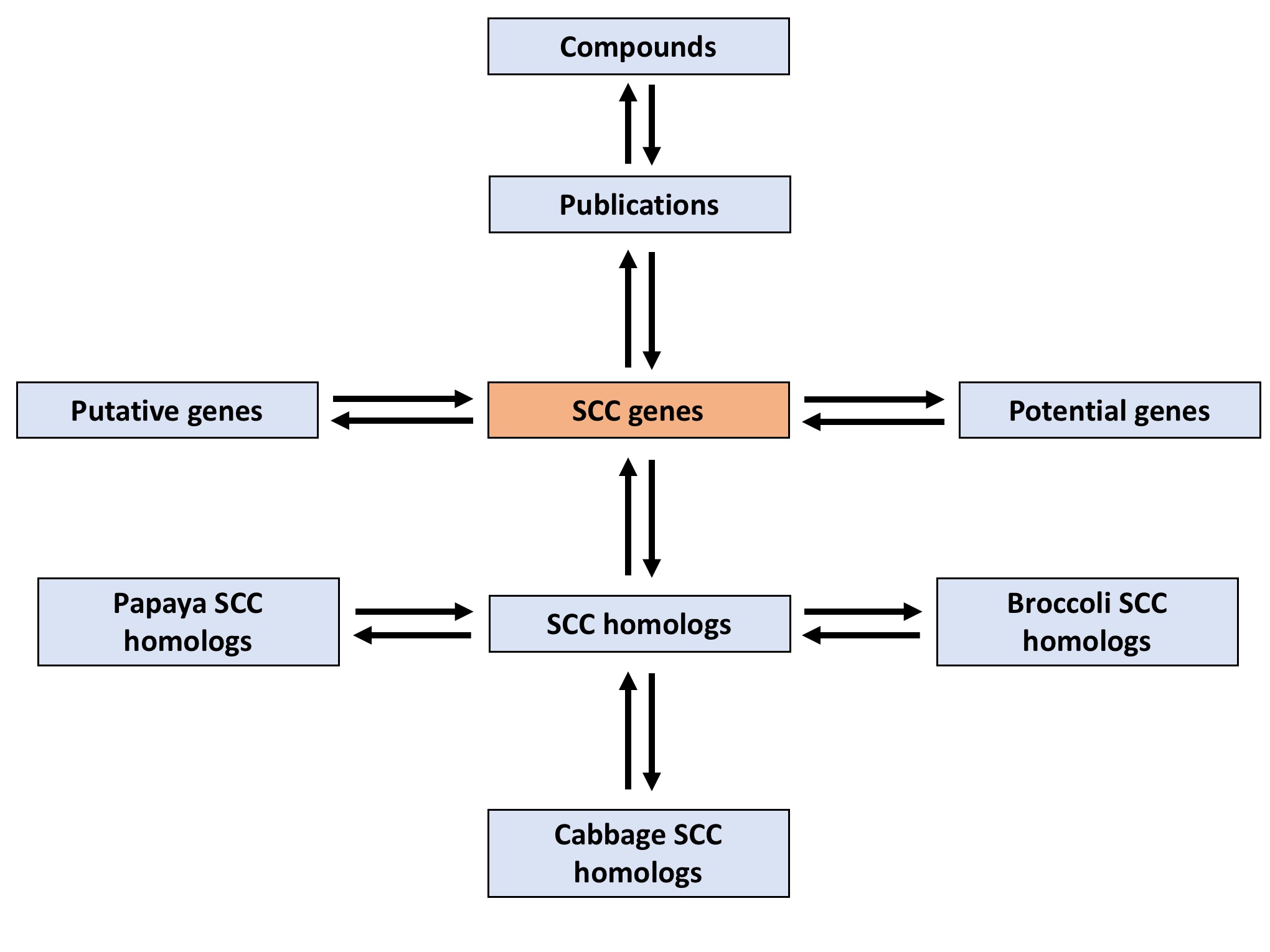
Figure 1 shows the integration of the tables entry in SuCComBase. The main table of SuCComBase is SCC genes/known Arabidopsis SCC genes table, which 147 genes are recorded. The blue-coloured boxes represent the tables that described SCC genes entity (e.g: Compounds, Publications, SCC homologs, etc).
SuCComBase can be used to search for genes, compounds, publications and homologs ID to match a keyword. User can also search the specific ID or keyword using the search box.
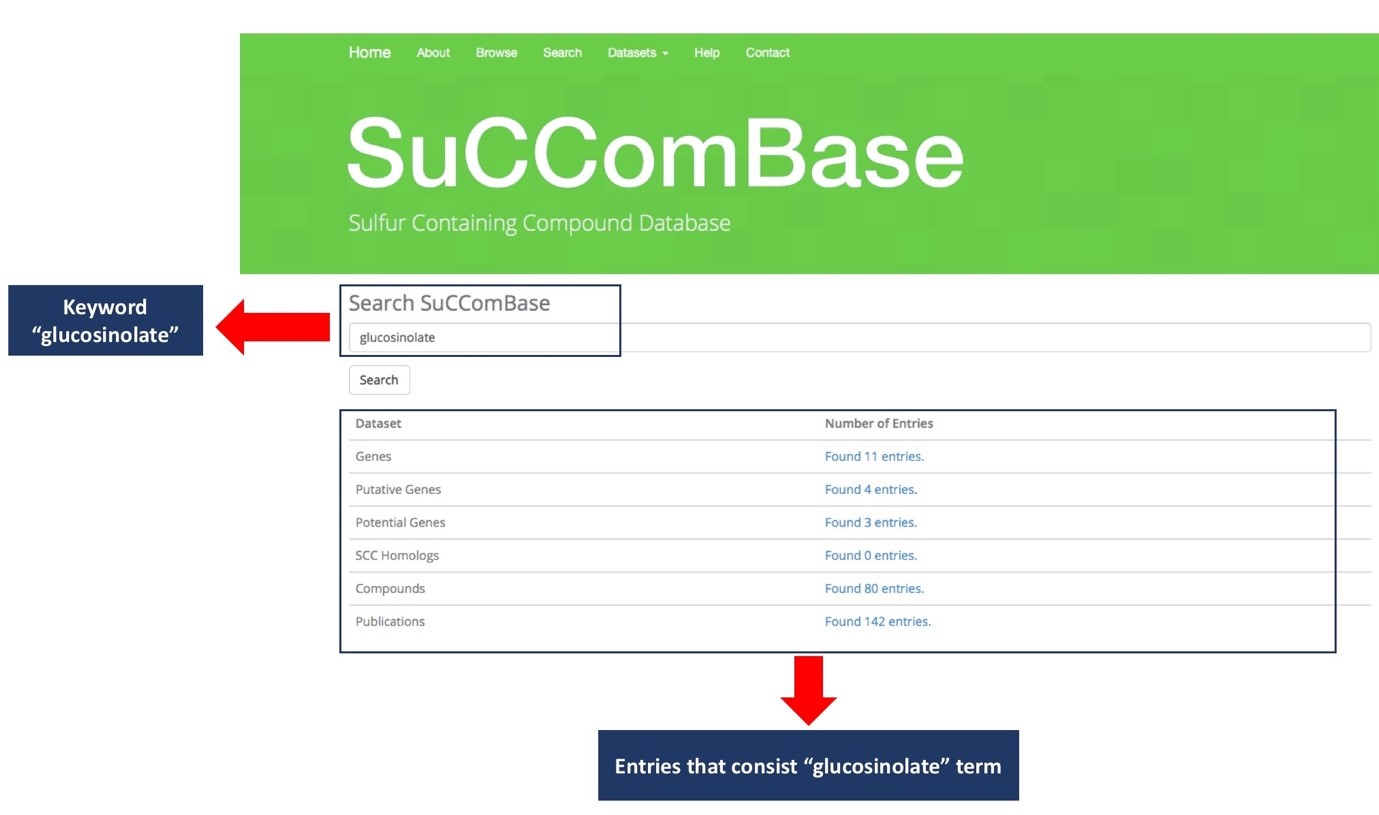
Figure 2 shows the page of SuCComBase with the ‘glucosinolate’ keyword as input in the search box. All entries in the datasets (genes, putative genes, potential genes, compounds, etc) will be shown once “glucosinolate” keyword is searched.
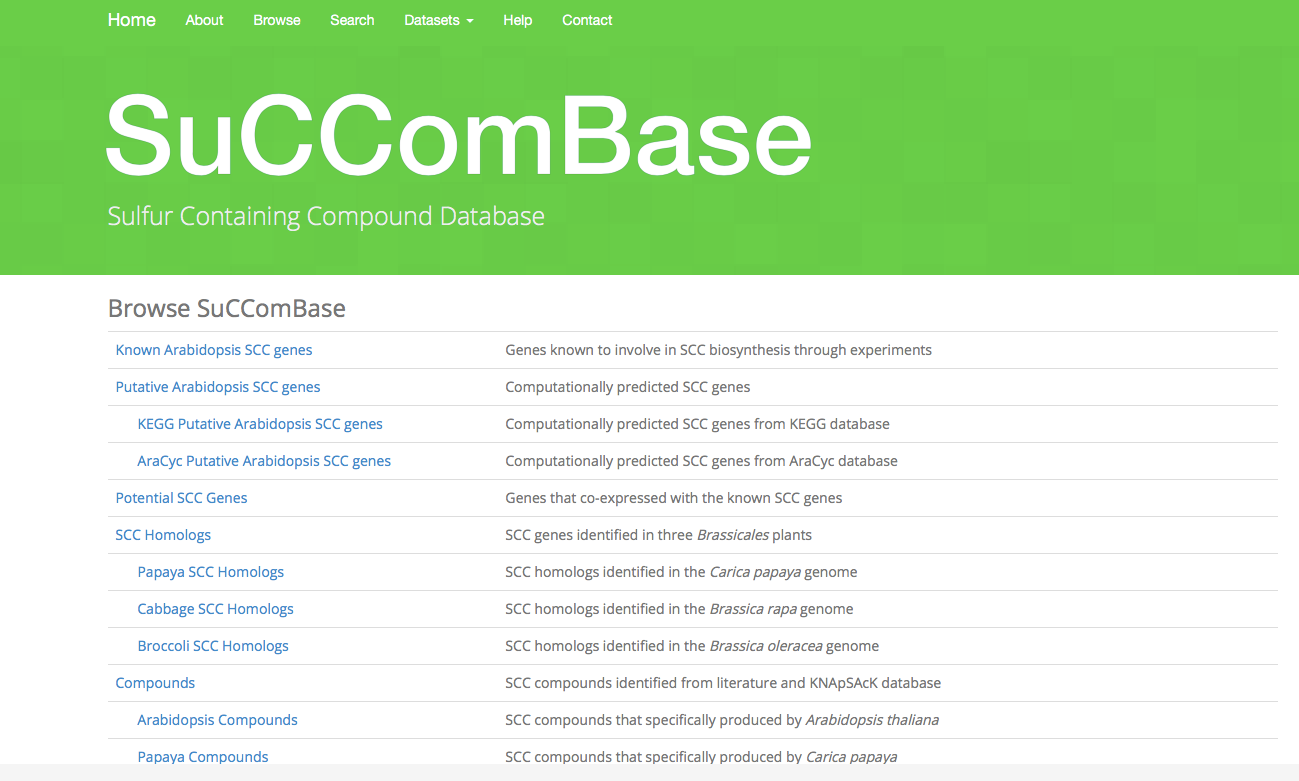
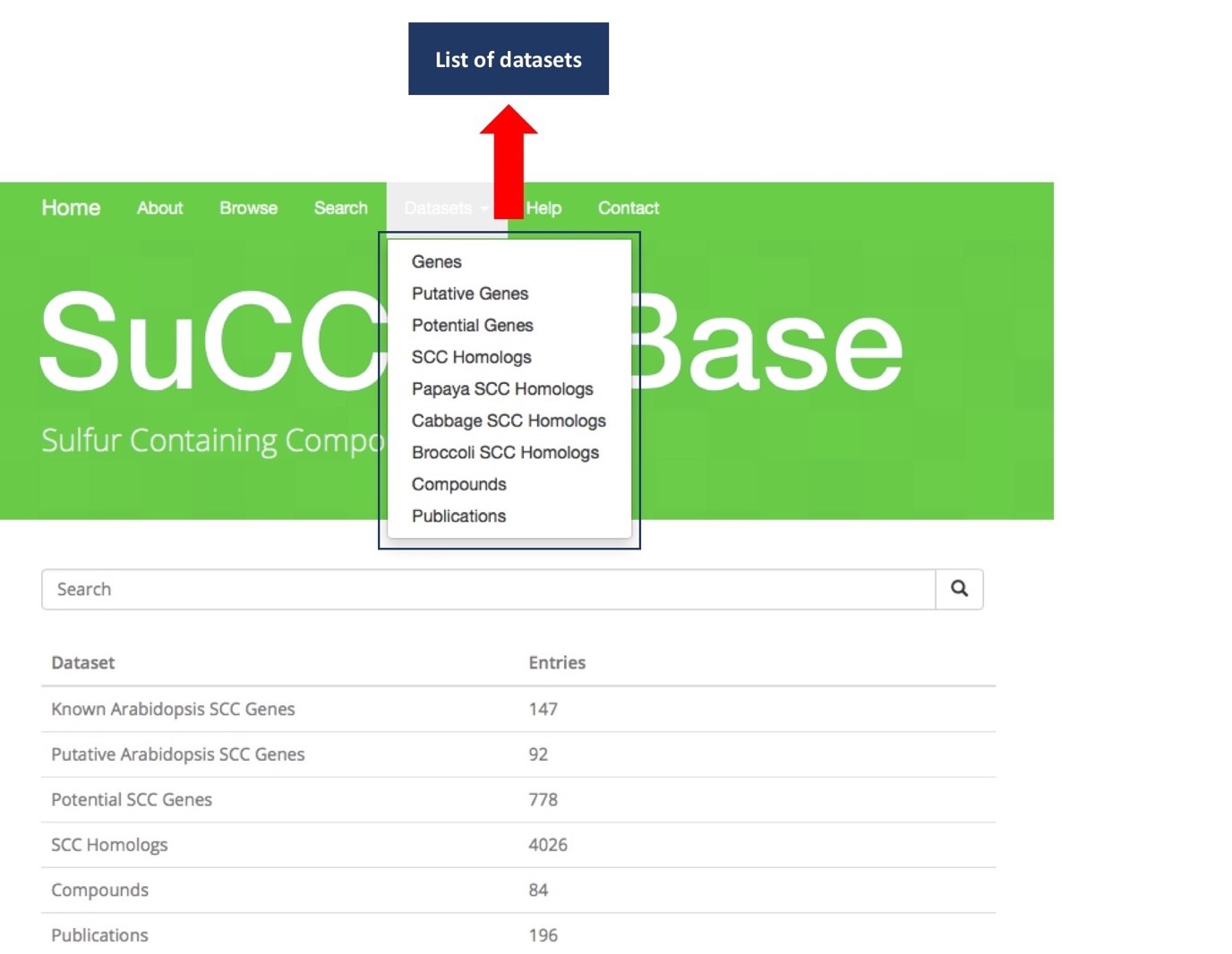
Browse page shows list of dataset tables for SuCComBase, for example. Known Arabidopsis SCC Genes, Putative Arabidopsis SCC Genes, Potential SCC Genes, SCC Homologs, Compounds and Publications. The description for every dataset was included.
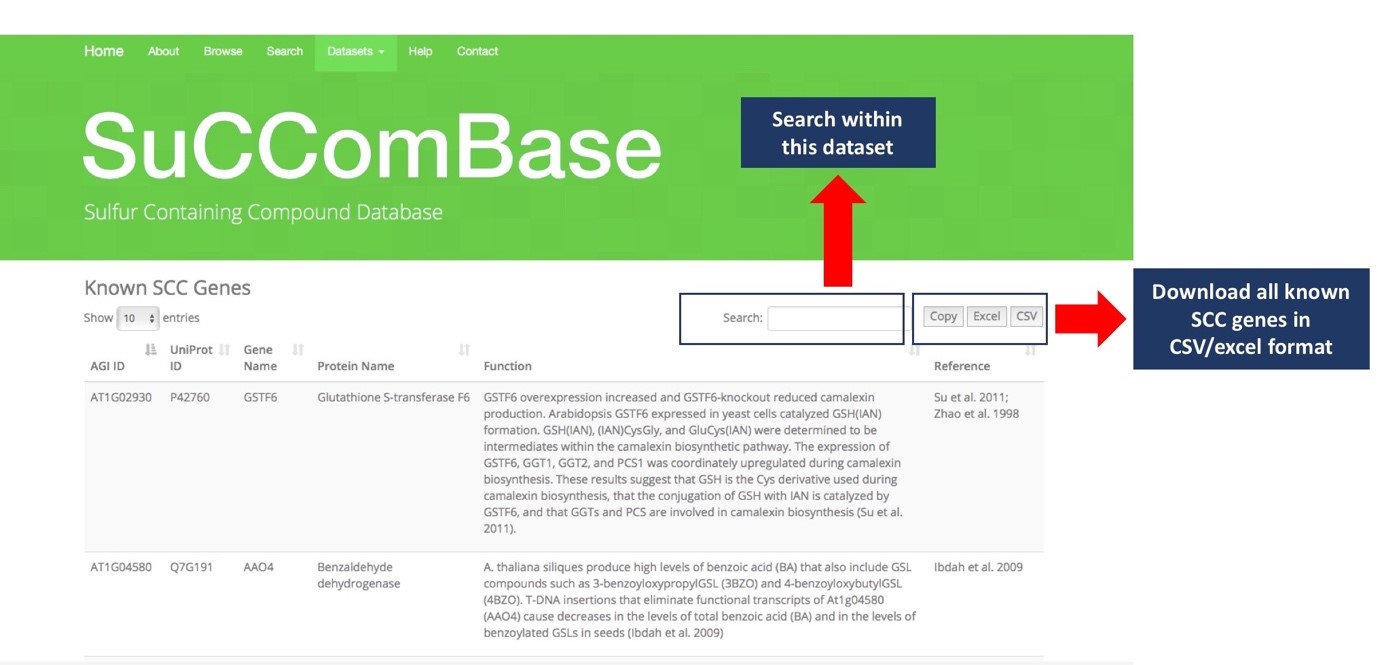
When you click on any of the datasets, a list of datasets will appear as in Figure 4. User can also search the gene of interest within the dataset and download all the information of Known SCC genes in .CSV or .xls format.
Datasets tab consists all datasets of SuCComBase. Datasets will redirect user to the target dataset pages by clicking on any of datasets.
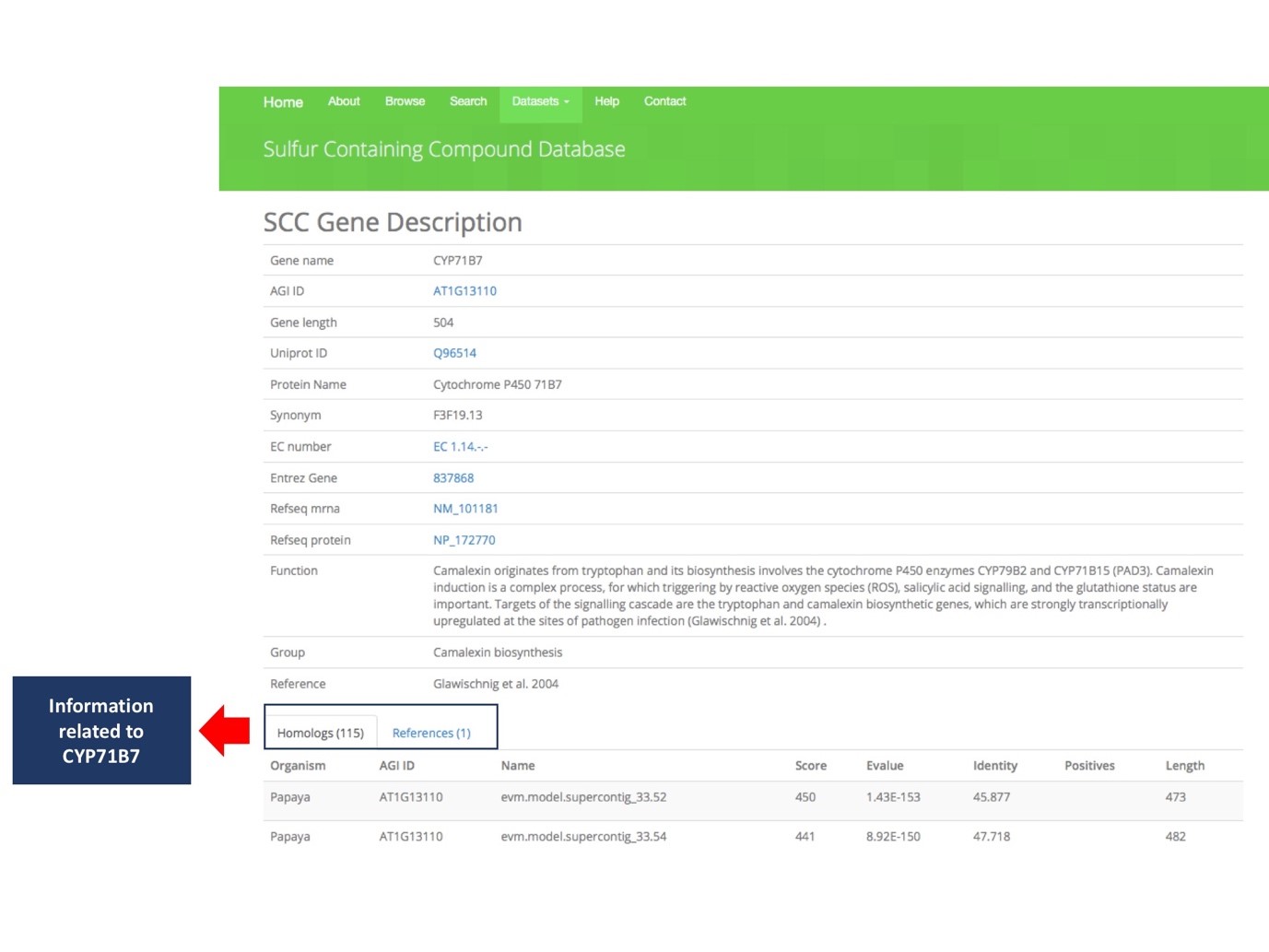
This segment showed the attributes provided when user click on any of the table from the Figure 4. The user will redirect to the SCC Gene Description as shown in Figure 6. All information including Gene name, AGI ID, Uniprot ID, Gene Function, Group of Biosynthesis, Homologs Gene, etc. Each tab of Homologs and References are clickable and contain sequence homology information in other Brassicales plant genomes. The example shows there are 25 genes are homolog to CYP71B7 and one reference found.